XTB Nano-Reactor for Essential Organic Reactions
The first cold day came to Kobe yesterday, with the temperature dropping to 0°C at night time. The red leaves of fall always linger into December, leading to a nice long fall, but winter is starting to creep up.

We have a new paper out now called XTB Nano-Reactor for Essential Organic Reactions. This is another collaboration with Nagasaki University. The purpose of this work was to investigate a low-cost protocol for discovering reaction mechanisms. A "Nano-Reactor" speeds up the search for reaction paths by performing high-temperature and pressure metadynamics simulations of the species of interest. By having your molecules bump into each other regularly with a lot of kinetic energy, you can force reactions to happen that would be outside of the timescales we can simulate with ab-initio methods at standard conditions. The nano-reactor has been implemented into the XTB program for use with low-cost semi-empirical quantum chemistry methods. Our paper finds that combination works great for a wide variety of organic reactions.
Now maybe that sounds exciting to you, but don't start reading the manual just yet. I'm sure you've heard all about Large Language Models (LLMs), and have been trying out things like ChatGPT yourself. Maybe during your interactions, you've found that the model not only can answer with text, but even do web searches, run scripts, produce images, and more. This is made possible by "tool calling". Essentially, the program running the model watches the output for a special syntax. If that appears, it uses it to run a script for the model, and returns the output to the model as further input.
The easiest way to make this work for your own custom tools is through the Model Context Protocol. This protocol makes it very easy to define a tool to be used by the model and connect it to a standard client. On the user side, you can also imagine building a whole library of tools for your model of choice to access. In fact, other scientists at RIKEN are working to make this even easier, with the dream of building a whole scientific ecosystem of tools.
Excited by this prospect, we decided to release an MCP server for the XTB Nano-Reactor alongside our paper. This server exposes two tool calls to an LLM: fetch an xyz file from pubchem and run the nano-reactor. Essentially, you can tell the model what system you want to simulate and under what conditions (or just leave that determination to the model!). The server automatically processes the output to extract reaction intermediates for the LLM to analyze.
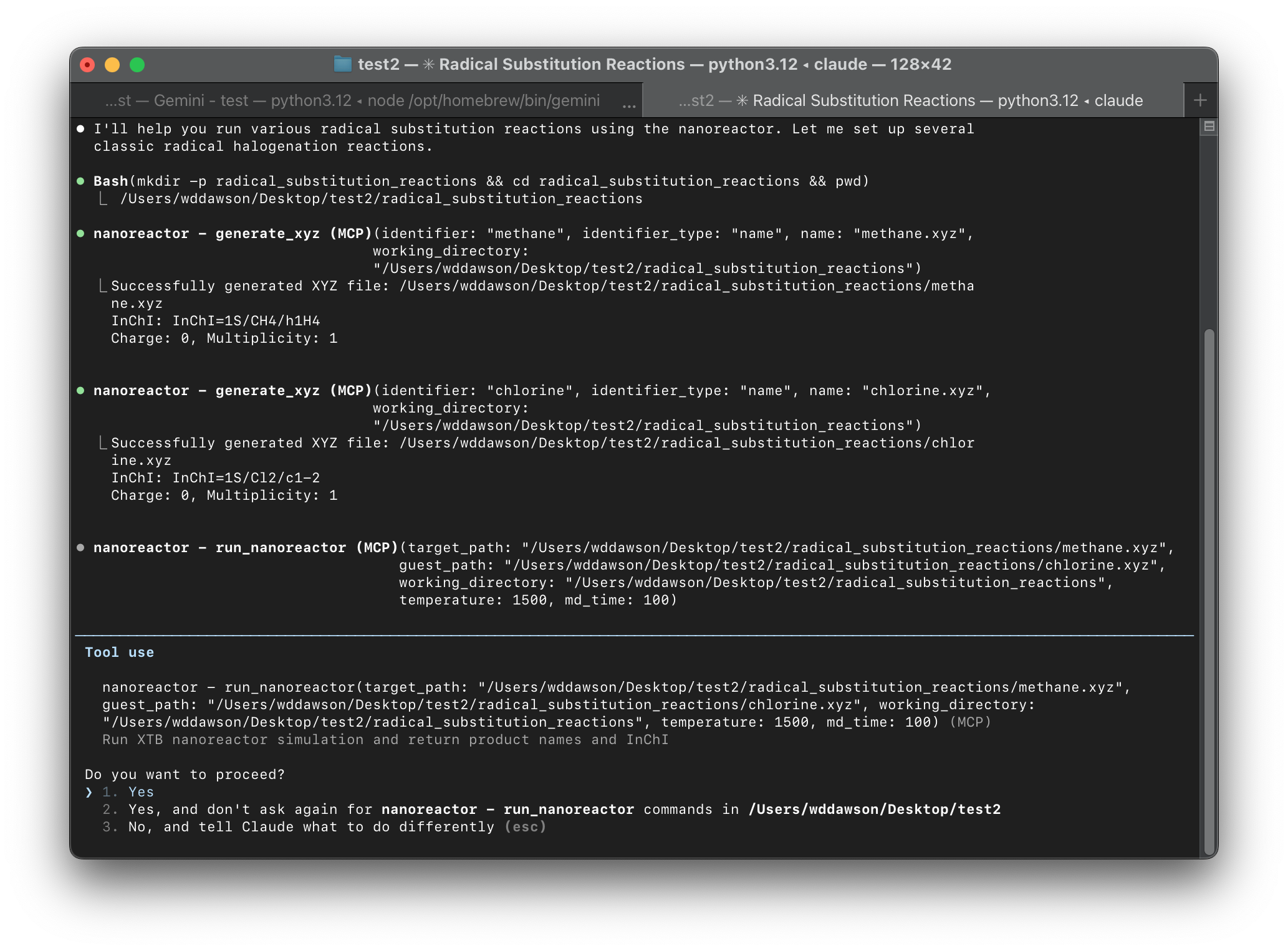
I've been using claude code as the front end to interact with the server. Some other front ends look nice these days but definitely need refinement. First install the mcp-reactor code and all its dependencies (openbabel, packmol, xtb), and then you can setup the code to work by typing: claude mcp add nanoreactor mcp-reactor. You might also want to modify the timeout, which recently seems to have been shortened.
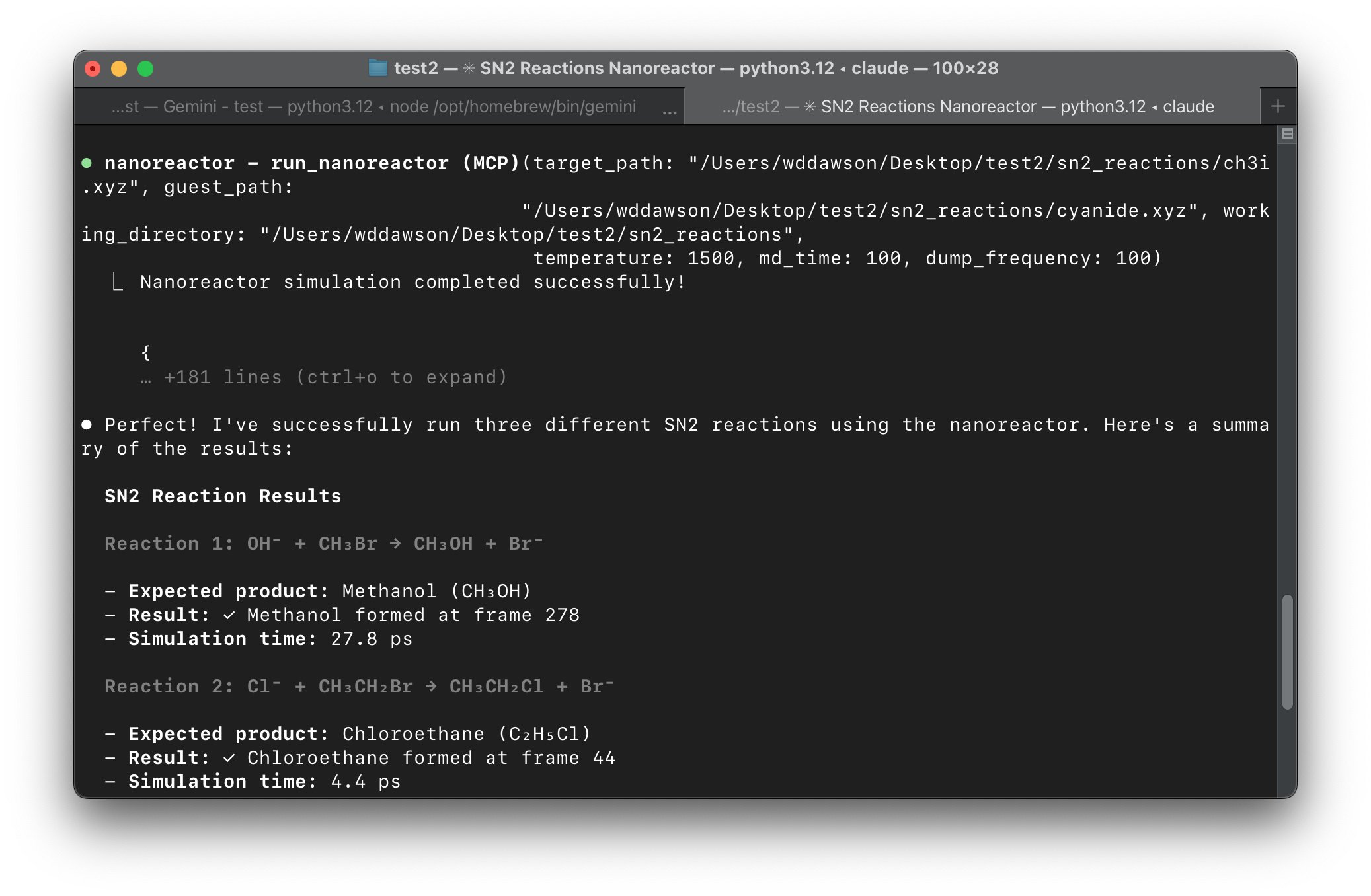
Our MCP server is at best a nice user interface - and at worse a toy. It's a very rigid workflow, and surely you'll find some exceptions that don't work. So where is the value added? I think we need to imagine a scenario where the LLM makes this tool call without you planning on it. Once we have a rich enough MCP ecosystem, maybe during a completely different study the LLM will realize running some nano-reactor simulations would help. So try encapsulating some of your tools this way, and let's see what we discover.